Examples
2D Fragmentation Example
This example demonstrates how to use Crispy to load a set of .agdd files obtained from a GranOO simulation of 2D disc impact and visualize fragmentation over time.
Code Example
import crispy as cp
detector = cp.FragmentDetector("path/to/your/agdd/files")
detector.build_fragments()
for it in range(detector.iterations_nb):
detector.plot2D(iteration=it)
This will generate one 2D image per iteration in your working directory, as well as a stack plot of the area repartition over iterations and a graph of the fragments heredity.
Note
Make sure the work directory contains valid .agdd files, the iteration sorting process is based on filename. For .agdd format, see details in the Quick Start section.
Preview of Generated Images
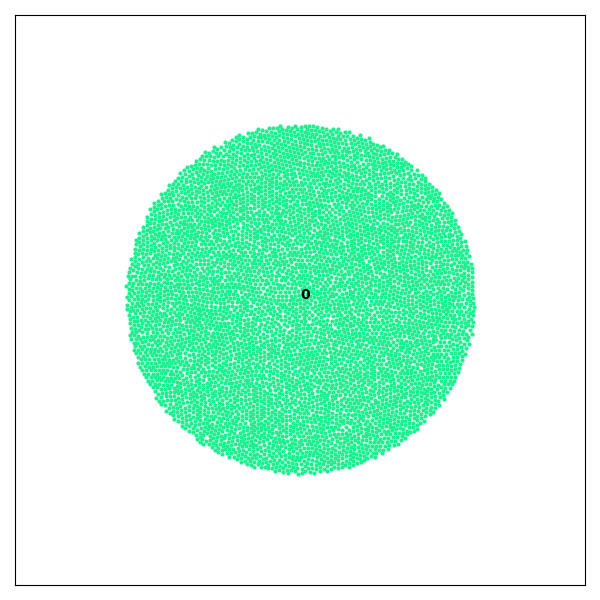

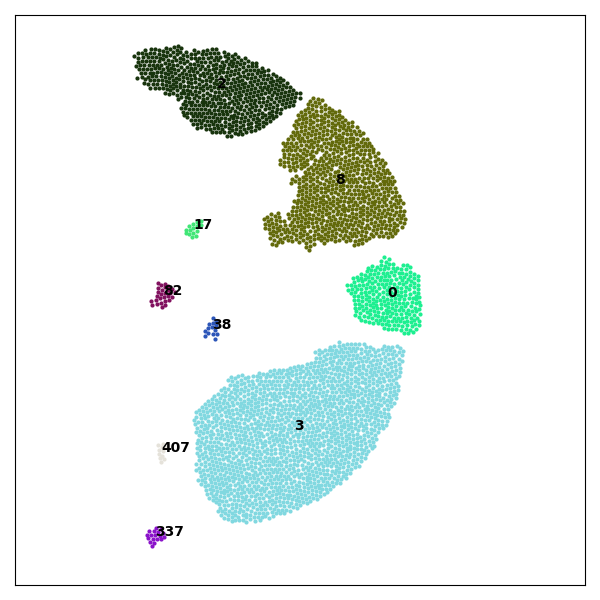
Run the Example
The full 2D fragmentation example is available in the project repository, inside the examples/2D_impact folder.
To run it locally, use the following commands:
cd examples/2D_impact
python 2D_impact.py
This script will execute the 2D fragmentation detection and save images for each iteration of the simulation in a subfolder examples/2D_impact/agdd/img of the working directory.
Note
Make sure you have installed all required dependencies listed in the Installation section before running the example.
You can inspect or modify the source code here:
examples/2D_impact/2D_impact.py.agdd input files are stored in
examples/2D_impact/agdd/
For more details on .agdd input file format, see the Quick Start section.